Exploring Phylogenetic Trees (with MEGA 4.0)
Developed by the UWW Bioinformatics Group and
the NSF sponsored National Evolutionary Synthesis Center
Welcome!
This site contains some useful definitions, resources, and links to help you become a skillful and informed user of phylogenetic trees.
It will serve as a handy reference tool as you produce your own make and interpret your own phylogenetic trees using MEGA 4.0.
Phylogenetic trees are extremely handy tools used by biologists to understand:
- the composition of genomes,
- relationships among genes in humans and other species,
- the functions of proteins that run our cells,
- historical relationships among diverse species,
- the processes that generate unique body shapes, and
- the origins of remarkable abilities in living organisms,
If you are curious about any of these topics, learning about phylogenetic trees will empower you to explore them in depth.
Useful Definitions:
Here are a few essential terms to help familiarize you with the making and reading of phylogenetic trees:
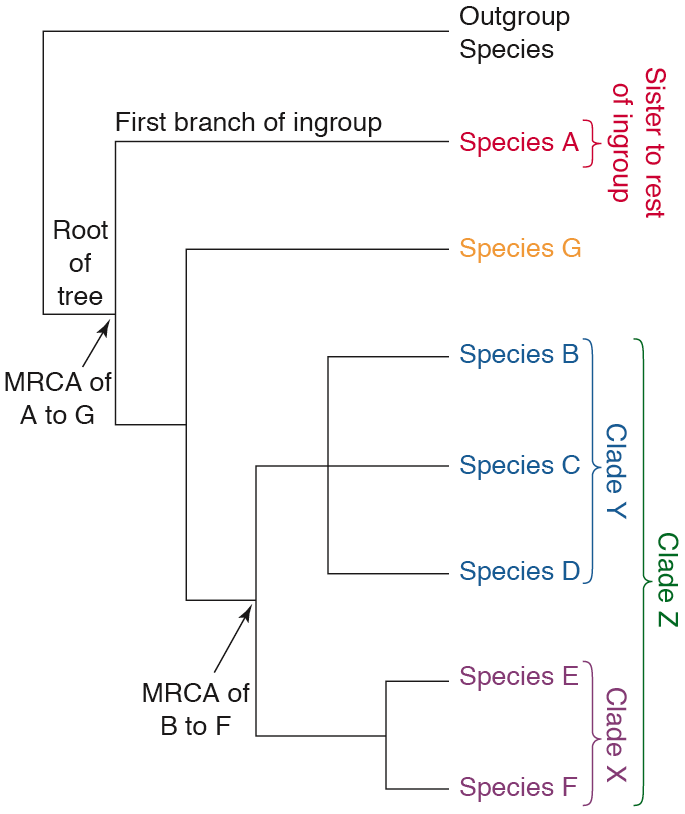
Phylogenetic tree (also called a “tree”) = a diagram that illustrates the evolutionary relationships among species, genes, or proteins. It is made of branches, nodes, terminals, and a root (see Figure below)
Terminal = a taxonomic group (like a species), gene, or protein that is included in a phylogenetic analysis (in the figure above, species A through G and the outgroup species are all terminals. These usually represent living species, genes, or proteins that are still around today.
Branches = the lines in a phylogenetic tree that connect terminals to nodes or one node to another
Nodes = points in a tree where branches intersect; they the represent ancestors of all species, genes, or proteins that descend from them. In the tree above, the branches that lead to Species E and Species F intersect at the node that represents their most recent common ancestor.
MRCA (most recent common ancestor) = the latest ancestor shared by a group of terminals; in the figure above, nodes in the tree are MRCAs of all terminals connected to them by branches to the right
Clades = natural groups of organisms, genes, or proteins in a phylogenetic tree, in the tree above, these are any MRCA and all of the terminals that descend from it
Root = the MRCA of the ingroup (point of attachment of the branch that connects the ingroup and the outgroup
Character = a feature common to all terminals that is sampled (e.g., eye color; base # 432 in a gene)
Character state = what a character looks like in an individual (e.g., eyes are blue; base # 432 is an A)
Useful Resources:
To download MEGA 4.0? Go here < http://www.megasoftware.net/>
For a step-by-step guide to building and interpreting phylogenetic trees, click here: Project_MEGA_Protocol.
For a starter file for the “How old are the parts of your body” project, click here: Project_MEGA_4.mas [Special note: use MEGA to open this or it will open as an Access file!]
Lesson plan for using MEGA to explore the evolution of human physical characteristics.
Kuzoff, R.K., Kemmeter, S.B., McKinnon, J.S., Thompson, C.P. 2009. Phylogenetic Analysis: How Old are the Parts of Your Body? Evolution: Education and Outreach, Vol 2(3):405-414. DOI 10.1007/s12052-009-0157-9
Useful Links:
For an excellent video introduction (10 min) to phylogenies and their uses, click here: http://www.peabody.yale.edu/exhibits/treeoflife/film_discovering.html
For a fun, interactive website (with 8 slides) that shows you how to interpret a phylogenetic tree, click here: http://www.pbs.org/wgbh/evolution/change/family/index.html
For a website (with 11 slides) that introduces terms and concepts used in reading and discussing phylogenetic trees, click here: http://evolution.berkeley.edu/evolibrary/article/0_0_0/evo_03
Questions, comments, suggestions, and the like:
Please send your questions and comments to: “MEGA Buddy” at nabt.mega@gmail.com